In late 2023, the first drug with the potential to slow the progression of Alzheimer’s disease was approved by the U.S. Food and Drug Administration (FDA). Alzheimer’s disease is one of several debilitating neurological diseases that affect one in eight people worldwide, and while this new drug is a step in the right direction, there is still a long way to go before we fully understand Alzheimer’s and other similar diseases.
“Reconstructing the complexity of how the human brain functions at the cellular level is one of the greatest challenges in neuroscience,” says Lars Jestevi, a technical staff member and algorithm developer in the Human Health and Performance Systems Group at MIT Lincoln Laboratory. “High-resolution, networked brain atlases could help us better understand disorders by pinpointing differences between healthy and diseased brains. However, progress has been hampered by a lack of tools to visualize and process very large brain imaging datasets.”
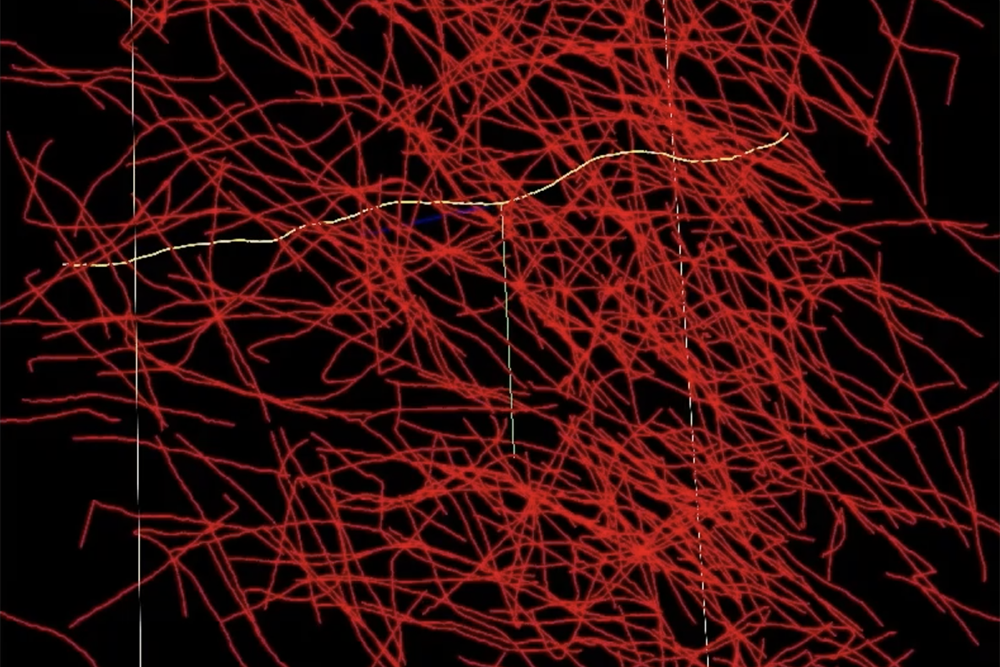
A networked brain atlas is essentially a detailed map of the brain that can help connect structural information to neural function. Building such an atlas requires processing and annotating brain imaging data. For example, the thin fibers that connect each axon or neuron must be traced, measured, and labeled with information. Current brain imaging data processing methods, such as desktop-based software or hand-oriented tools, are not yet designed to handle human brain-scale data sets. As a result, researchers often spend a lot of time wading through oceans of raw data.
Gjesteby leads a project to build NeuroTrALE (Neuron Tracing and Active Learning Environment), a software pipeline that brings machine learning, supercomputing, and ease of use and accessibility to this brain mapping task. NeuroTrALE automates most of the data processing and presents the output in an interactive interface that allows researchers to edit and manipulate the data to highlight, filter, and search for specific patterns.
Unraveling the thread
One of the features of NeuroTrALE is its use of a machine learning technique called active learning. NeuroTrALE’s algorithms are trained to automatically label incoming data based on existing brain imaging data, but unfamiliar data can introduce potential errors. Active learning allows users to manually correct errors, teaching the algorithm to improve the next time it encounters similar data. Combining automation and manual labeling ensures accurate data processing while reducing the burden on the user.
“Imagine taking an X-ray of a ball of thread. You’d see all these intersecting, overlapping lines,” says Michael Snyder of the lab’s Homeland Decision Support Systems Group. “When two lines cross, is one piece of thread bending 90 degrees, or is one going straight up and the other going straight up? With NeuroTrALE’s active learning, a user can trace that strand of thread once or twice and train the algorithm to follow it correctly going forward. Without NeuroTrALE, you would have to trace the ball of thread—or in this case, the axons of the human brain—every time.” Snyder works with David Chavez, a software developer and associate on the NeuroTrALE team.
NeuroTrALE offloads most of the labeling burden from the user, allowing researchers to process more data, faster. The axon tracing algorithm also leverages parallel computing to distribute computations across multiple GPUs simultaneously, enabling faster and more scalable processing. Using NeuroTrALE, the team reduced the computing time required to process 32 gigabytes of data by 90 percent compared to existing AI methods.
The team also showed that processing times do not increase in proportion as data volumes increase significantly. For example, in a recent study, they showed that a 10,000% increase in data set size resulted in a 9% and 22% increase in total data processing time using two types of central processing units, respectively.
“With an estimated 86 billion neurons forming 100 trillion connections in the human brain, manually labeling all the axons in a single brain would take a lifetime,” adds Benjamin Roop, one of the algorithm developers on the project. “This tool has the potential to automate connectome generation for single individuals, and even many individuals, opening the way to studying brain diseases at a population level.”
The path to open source discovery
The NeuroTrALE project was an internally funded collaboration between Lincoln Laboratory and Professor Kwanghun Chung’s lab on the MIT campus. The Lincoln Lab team needed to build a way for Chung Lab researchers to analyze and extract useful information from the vast amounts of brain imaging data flowing into the MIT SuperCloud, a supercomputer operated by Lincoln Laboratory to support MIT research. Lincoln Lab’s expertise in high-performance computing, image processing, and artificial intelligence made it ideally suited to tackle this challenge.
In 2020, the team uploaded NeuroTrALE to SuperCloud, and in 2022, the Chung Lab produced the results. In one study, scienceThey used NeuroTrALE to quantify prefrontal cortex cell density in relation to Alzheimer’s disease. Brains with Alzheimer’s disease had lower cell density in certain areas than brains without Alzheimer’s disease. The same team also found which parts of the brain tended to have harmful neurofibrillary tangles in Alzheimer’s brain tissue.
Work on NeuroTrALE continues with funding from Lincoln Laboratory and National Institutes of Health (NIH) to build out the capabilities of NeuroTrALE. The user interface tools are currently being integrated with Google’s Neuroglancer program, an open-source, web-based viewer application for neuroscience data. NeuroTrALE adds the ability for users to dynamically visualize and edit annotated data, and allows multiple users to work on the same data simultaneously. Users can create and edit multiple shapes, such as polygons, points, and lines, to facilitate annotation, and can even customize the color scheme for each annotation to distinguish neurons in dense areas.
“NeuroTrALE provides a platform-independent, end-to-end solution that can be easily and quickly deployed via containers to standalone, virtual, cloud, and high-performance computing environments,” said Adam Michaleas, a high-performance computing engineer in the lab’s AI Technologies Group. “Furthermore, it significantly improves the end-user experience by providing real-time collaboration capabilities within the neuroscience community through data visualization and concurrent content review.”
In keeping with the NIH’s mission to share research products, the team’s goal is to make NeuroTrALE a completely open-source tool that anyone can use. And Gjesteby says these types of tools are necessary to reach the ultimate goal of mapping the entire human brain for research and ultimately developing drugs. “This is a grassroots community effort to make sure that data and algorithms are shared and accessible to everyone.”
Codebase for Axon tracing, Data Managementand Interactive User Interface All features of NeuroTrALE are publicly available under an open source license. Contact us. Las Jestevi More information about using NeuroTrALE
